HepNet™ is a proprietary, hepatocyte-specific, computational biology platform that enables:
- Mining of 100s of integrated data sources to distil new mechanistic knowledge of hepatocyte biology
- Application of sophisticated network biology analytics
- The identification of novel target genes to unlock differentiated therapeutic development at scale
- Increased automation of the early discovery stages
Highly predictive siRNA construct design building on carefully curated proprietary and public data sources, HepNet™ generates and analyses biological network models, providing a novel and mechanistic approach to target identification.
HepNet™ addresses key limitations of the biopharmaceutical industry, including extracting value from big data, addressing complex diseases and improving translatability in R&D. It focuses on tackling the hardest challenge, which is often being overlooked: human biology.
We focus on tackling the hardest challenge, which is being overlooked: human biology.
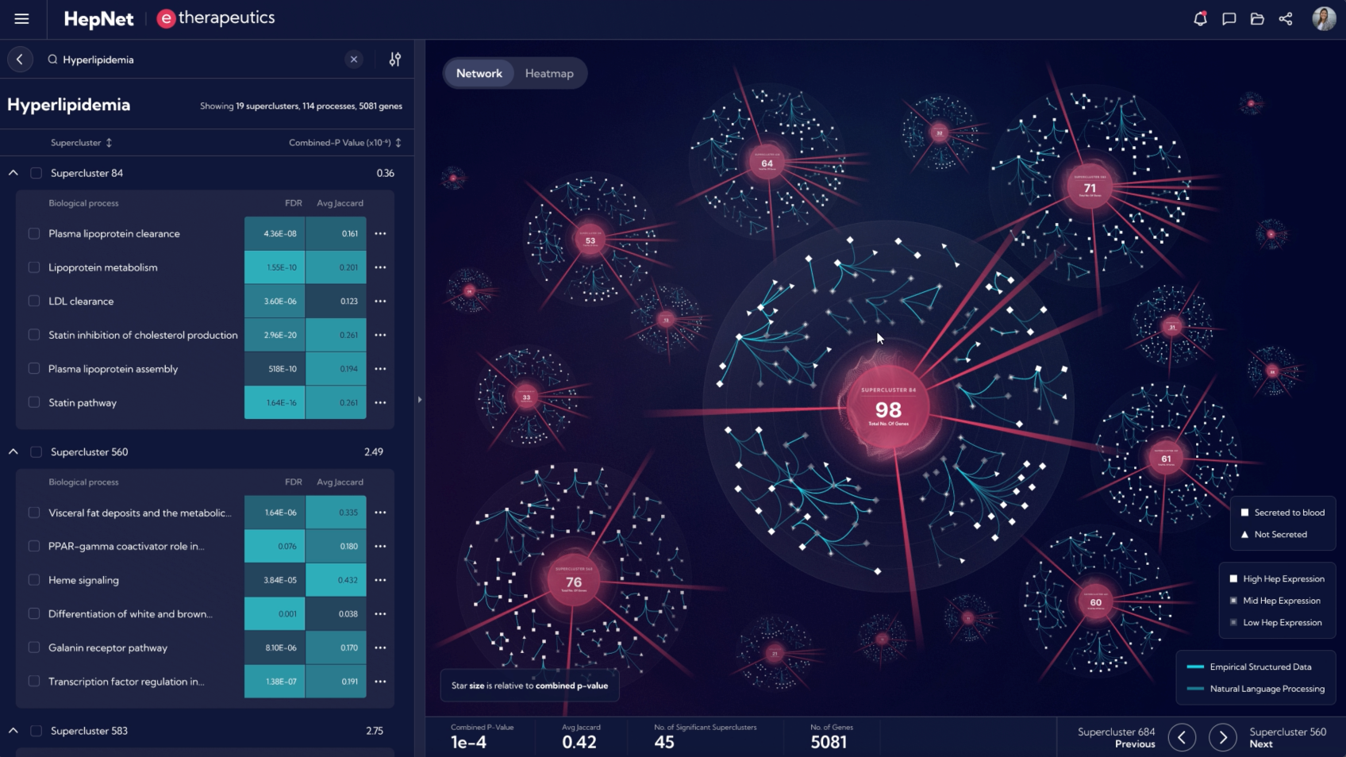
Target identification with HepNetTM
HepNet™ rapidly identifies targets by:
- Generating in silico network models that help understand and analyse connections between genes and proteins in a hepatocyte-centric biological ecosystem.
- Interrogating biological processes to generate hypotheses and test millions of potential interventions including the downstream effects of silencing a specific gene.
- Uncovering novel biology and the right mechanisms/nodes to target as a therapeutic strategy in a given indication.
This computational network biology approach not only identifies high-quality targets but helps to better understand complex systems biology supporting the mechanism of action and makes the whole target identification process reproducible end-to-end so that the discovery of each novel drug is not a brand-new scientific problem.
Working with disease-driving biological networks instead of pathways provides a novel approach to drug discovery that explicitly considers the true complexity of biology. By representing relevant biological systems more holistically, HepNet™ increases the likelihood of identifying target genes with a robust mechanism of action, supporting the development of effective therapies with disease-modifying potential.
HepNetTM as a dynamic knowledge resource
HepNet™ includes a combination of large-scale public, licensed, and proprietary data, which is integrated and curated using AI and network-aware methods. It makes sense of complex datasets, creating actionable insights by placing genes in the context of the biological networks they belong to. The novelty of this biology-centred approach, underpinned by the integration of multiple disparate data sets with our hepatocyte-specific interactome, enables the characterisation of currently undruggable disease processes and the identification of the best network-aware intervention strategies.
We have built a proprietary hepatocyte-focussed Knowledge Graph that is further enhanced with data derived from Natural Language Processing (NLP) and AI-driven predictive approaches to knowledge inference. This means we can discover non-obvious and hidden relationships in data and have the capabilities to impute missing information.
The recent revolution in large language models (LLMs), including GPT-4, has enabled us to significantly increase our ability to ingest and process text, leverage unstructured data at scale, and derive tangible knowledge from buried data that might already be in the public domain.
HepNet™ will continue to evolve as we expand the variety of information inputs. By placing LLMs at the core of our computation, and harnessing GPT-4’s capabilities, we can now create specialised LLM “agents” which will transform HepNetTM into a dynamic knowledge resource. GPT-4 and LLM integration will provide a unifying framework from which to drive every aspect of our pipeline, and positioning e-therapeutics as a world leader in hepatocyte biology and related diseases.
Our long-term vision is to fully automate the preclinical drug discovery process- to accelerate the development of life-transforming treatments.
14M
hepatocyte specific data points
16,000
genes
1,300
hepatocyte associated diseases in knowledgebase
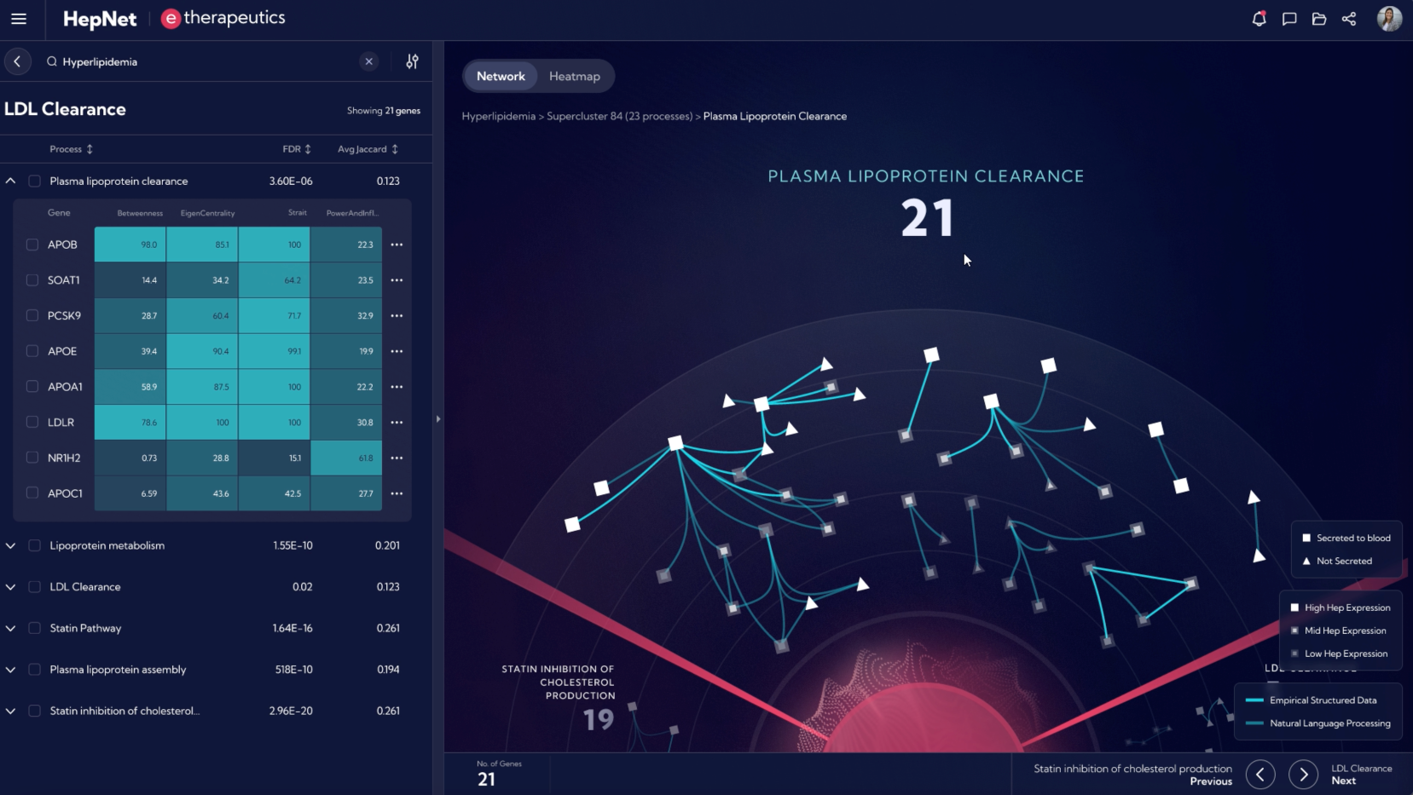
Images from the HepNetTM computational platform are for illustrative purposes only